.
Condición fitosanitaria: Presente ampliamente distribuida
Grupo de cultivos: Oleaginosas
Especie hospedante: Soja (Glycine max)
Rango de hospedantes: específico / estrecho. La soja es el único hospedante económicamente importante. También se han reportado varias especies de lupinos (Lupinus spp.) como hospedantes (Tyler, 2006).
Epidemiología: monocíclica, subaguda.
Etiología: Pseudohongo. Necrotrófico (breve fase inicial hemibiotrófica entre 24 a 36 hpi)
Agente causal: Phytophthora sojae Kauffmann & Gerdemann
Taxonomía: Eukaryota > Stramenopiles > Oomycetes > Peronosporales > Peronosporaceae > Phytophthora
.
.
.
Antecedentes
Esta enfermedad fue una de las primeras patologías detectadas en el cultivo de soja, luego de la podredumbre de tallos y vainas por Sclerotinia sclerotiorum y la mancha marrón por Septoria glycines.
.
Síntomas
El síntoma típico de la enfermedad está constituido por la decoloración parda del tallo desde la superficie del suelo que contrasta con los tejidos verdes superiores. La pudrición de raíces y de la base del tallo conduce finalmente al marchitamiento de las plantas. El patógeno infecta en cualquier etapa del cultivo. Puede causar tizón en pre- y post-emergencia (ver tizón de plántulas). Los síntomas generales, incluyen amarillamiento de hojas y marchitamiento de las plantas. Los síntomas en plantas adultas antes del marchitamiento consisten en el oscurecimiento de la base del tallo que avanza por las ramificaciones hasta el quinto o sexto entrenudo y contrasta con los tejidos verdes superiores aún sanos. Las raíces y el tejido vascular son invadidos por el patógeno. Finalmente se produce la muerte de la planta y las hojas quedan adheridas al tallo. Cultivares tolerantes pueden mostrar pudrición del sistema radical, pero sobreviven.
.
.
- Síntomas de la pudrición de la raíz por Phytophthora en plántulas de soja: (A) ligera decoloración de la raíz principal y las raicillas, con una lesión empapada de agua en el tallo; (B) decoloración de la raíz principal y de las raicillas de marrón a marrón oscuro, con una lesión de color chocolate en el tallo; (C1-C3) daño de hipocótilo y cotiledón; y (D) pudrición de semillas. Autor: Chang et al., 2017.
.
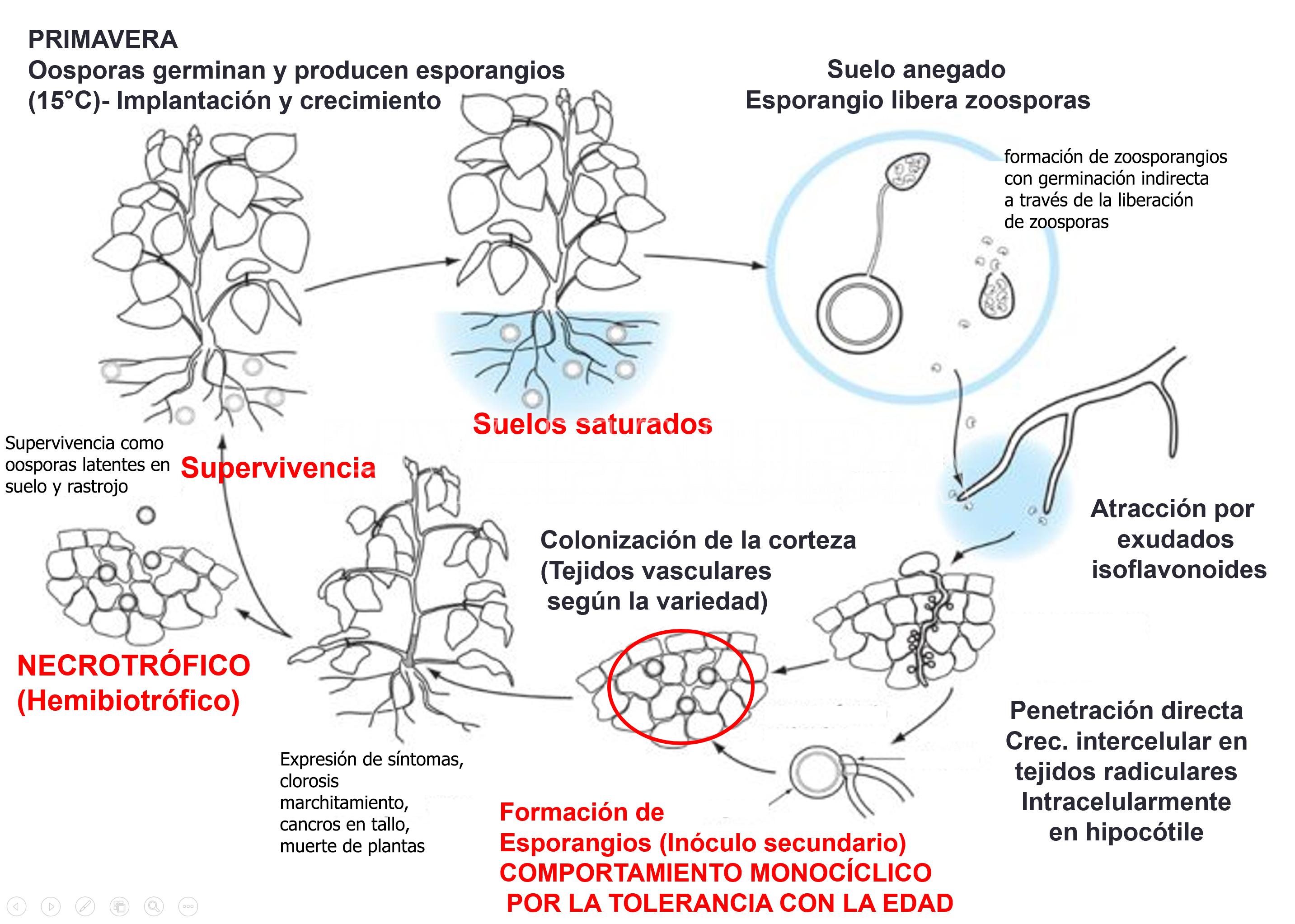
Condiciones ambientales predisponentes para el establecimiento de la enfermedad, Epidemiología
El patógeno es un oomycete, y por lo tanto forma esporas flageladas (zoosporas) que se dispersan fácilmente en medio acuoso. Este oomycete tiene la capacidad de persistir en el suelo y en los rastrojos como oosporas. Las oosporas permanecen latentes durante años, ya que las esporas producidas en la misma estación generalmente deben acumular un período de suelo frío antes de que ocurra la germinación.
Las condiciones ambientales predisponenetes son suelos húmedos y fríos, pobremente drenados, pesados (con altos contenidos de arcilla), presencia de rastrojo infectado y drenaje deficiente, predisponen al desarrollo de la enfermedad, la cual se manifiesta también con mayor intensidad en áreas bajas de los lotes. El desarrollo de epidemias se ve favorecido por la acumulación de agua en el suelo ya que facilita la dispersión de las zoosporas, por lo cual los daños son más severos en suelos compactos, en las cabeceras de los lotes y en años lluviosos. La presencia de rastrojos infestados del cultivo anterior aumenta la densidad de inóculo del patógeno ya que las oosporas quedan protegidas por los tejidos muertos.
En condiciones de suelo casi saturado, las oosporas preacondicionadas germinan para formar micelio que luego produce zoosporangios. Los zoosporangios maduran rápidamente y cuando el suelo se satura, liberan zoosporas para infectar las raíces del hospedante. Las zoosporas son negativamente geotácticas y son atraídas por exudados de raíces como daidzeina y genisteína (isofalvonoide). Una vez liberadas, las zoosporas nadan hacia las raíces de la planta hospedante. Una vez que entran en contacto con una raíz, las zoosporas se enquistan, pierden sus flagelos y forman una pared celular. Luego, germinan para formar un tubo germinativo y penetran en la epidermis de la raíz a través de la formación de un apresorio.
.
- Ciclo biológico-agronómico de la pudrición del tallo de soja por Phytophthora
.
Manejo de la enfermedad
P. sojae es un patógeno de alta especificidad, si bien la soja no es el único hospedante, no ataca otros cultivos extensivos de verano en la Argentina. La enfermedad es fácilmente controlable a través de la resistencia genética que es conferida por genes mayores Rps. Durante muchos años se observó baja variabilidad en virulencia ya que prevaleció la raza 1 (Barreto et al, 1995, 1998). Sin embargo, la liberación de cultivares resistentes indujo la aparición o incremento de frecuencia de nuevas razas lo cual dificulta el manejo de la enfermedad.
Desde hace varios años, se han encontrado fuentes de resistencia a P. sojae (Dorrance y Schmitthenner, 2000; Yang et al., 2020; Bolaños-Carriel et al., 2021). En las últimas décadas, se han mapeado unas pocas docenas de loci / alelos de resistencias (Rps) en varias regiones genómicas de la soja enriquecidas con genes que codifican proteínas receptoras de resistencia específicas (proteínas de sitio de unión a nucleótidos de repetición rica en leucina (NBS-LRR o NLR) en base al ensamblaje del genoma de referencia de soja v2.0 (soybase.org) (Sahoo et al., 2017; Zhong et al., 2018; Matthiesen et al., 2016; Jiang et al., 2020; Matthiesen et al., 2021). Sin embargo, debido a la gran variabilidad genética en las poblaciones del patógeno en constante evolución (Abney et al., 1997; Yang et al., 2019), muchos de esos loci / alelos se han vuelto ineficaces para los aislados del patógeno recientemente evolucionados y se han aislado y/o validado funcionalmente pocos loci/alelos de Rps (Dong et al., 2011; Yan y Nelson, 2019; Yang et al., 2019). Por lo tanto, la introgresión/piramidación efectiva de genes Rps específicos en cultivares de soja de élite para lograr una resistencia sostenida sigue siendo un desafío. Recientemente, Wang et al. (2021) encontraron que el gen de resistencia Rps11 confiere resistencia de amplio espectro a P. sojae; y Sahoo et al. (2021) encontraron que dos genes Rps ( Rps12 and Rps13) funcionales estrechamente ligados con una especificidad de raza distinta proporcionan una resistencia de amplio espectro en la soja.
Para el manejo de este patosistema es necesario integrar varias herramientas de manejo, como uso de cultivares resistentes, rotaciones (2 a 3 años con gramíneas) y tratamiento de semillas con metalaxil. Mejorar el drenaje si es posible. P. sojae tiene alta variabilidad patogénica (razas) actualmente en nuestro país, por lo cual los cultivares que resistentes a la raza 1, que es la más común, pueden no serlo para las que se encuentren en el lote afectado.
.
.
- Autor: Daren Mueller, Iowa State University
- Autor: Daren Mueller, Iowa State University
.
- Campo encharcado con P sojae. Autor: Dr. Pablo Grijalba.
- Manchón causado por P sojae. Autor: Dr. Pablo Grijalba.
- Manchón causado por P sojae. Autor: Dr. Pablo Grijalba.
- Manchón causado por P sojae. Autor: Dr. Pablo Grijalba.
- Planta muerta por P sojae. Autor: Dr. Pablo Grijalba.
- Planta muerta por P sojae. Autor: Dr. Pablo Grijalba.
- Planta muerta por P sojae. Autor: Dr. Pablo Grijalba.
- Autor: Tom Allen
- Autor: Tom Allen
- Autor: Tom Allen
- Autor: Tom Allen
.
.
.
Bibliografía
Abad ZG, Burgess T, Redford AJ, et al. (2022) IDphy: An international online resource for molecular and morphological identification of Phytophthora. Plant Disease. doi: 10.1094/PDIS-02-22-0448-FE
Abeysekara NS, Matthiesen RL, Cianzio SR, et al. (2016) Novel Sources of Partial Resistance against Phytophthora sojae in Soybean PI 399036. Crop Science 56: 2322–2335. doi: 10.2135/cropsci2015.09.0578
Abney TS, Melgar JC, Richards TL, et al. (1997) New Races of Phytophthora sojae with Rps1-d Virulence. Plant Disease 81(6): 653-655. doi: 10.1094/PDIS.1997.81.6.653
Adigun OA, Pham TH, Grapov D, et al. (2023) Phyto-oxylipin mediated plant immune response to colonization and infection in the soybean-Phytophthora sojae pathosystem. Front Plant Sci. 14: 1141823. doi: 10.3389/fpls.2023.1141823
Ai G, Si J, Cheng Y, et al. (2023) The oomycete-specific BAG subfamily maintains protein homeostasis and promotes pathogenicity in an atypical HSP70-independent manner. Cell Rep. 42(11): 113391. doi: 10.1016/j.celrep.2023.113391
Arsenault-Labrecque G, Santhanam P, Asselin Y, et al. (2022) RXLR effector gene Avr3a from Phytophthora sojae is recognized by Rps8 in soybean. Mol Plant Pathol. doi: 10.1111/mpp.13190
Avila-Quezada GD, Rai M (2023) Novel nanotechnological approaches for managing Phytophthora diseases of plants. Trends Plant Sci.: S1360-1385(23)00102-4. doi: 10.1016/j.tplants.2023.03.022
Belisle RJ, Hao W, Riley N, et al. (2022) Root absorption and limited mobility of mandipropamid as compared to oxathiapiprolin and mefenoxam after soil treatment of citrus plants for managing Phytophthora root rot. Plant Dis. doi: 10.1094/PDIS-07-22-1699-RE
Bolaños-Carriel C, Batnini A, McHale LK, Dorrance AE (2021) Identification of resistance loci towards phytophthora sojae (Rps) in South Korean soybean plant introductions 407974B and 424487B. Crop Sci. doi: 10.1002/csc2.20596
Brasier C, Scanu B, Cooke D, Jung T (2022) Phytophthora: an ancient, historic, biologically and structurally cohesive and evolutionarily successful generic concept in need of preservation. IMA Fungus 13(1): 12. doi: 10.1186/s43008-022-00097-z
Bronkhorst J, Kasteel M, van Veen S, et al. (2021) A slicing mechanism facilitates host entry by plant-pathogenic Phytophthora. Nat Microbiol 6: 1000–1006. doi: 10.1038/s41564-021-00919-7
Bronkhorst J, Kots K, de Jong D, et al. (2022) An actin mechanostat ensures hyphal tip sharpness in Phytophthora infestans to achieve host penetration. Sci Adv. 8(23): eabo0875. doi: 10.1126/sciadv.abo0875
, , , et al. (2020) Towards a best practice methodology for the detection of Phytophthora species in soils. Plant Pathology 00: 1– 11. doi: 10.1111/ppa.13312
, , , et al. (2021) Stepwise accumulation of mutations in CesA3 in Phytophthora sojae results in increasing resistance to CAA fungicides. Evol Appl. 14: 996– 1008. doi: 10.1111/eva.13176
Chang KF, Hwang SF, Ahmed HU, et al. (2017) First report of Phytophthora sojae causing root rot in soybean [Glycine max (L.) Merr.] in Alberta, Canada. Crop Protection 91: 49-56. doi: 10.1016/j.cropro.2016.09.006
Cao J, Qiu M, Ye W, Wang Y (2022) Phytophthora sojae Transformation Based on the CRISPR/Cas9 System. Bio Protoc. 12(6): e4352. doi: 10.21769/BioProtoc.4352
Cerritos Garcia DG, Huang SY, Kleczewski NM, Mideros S (2022) Virulence, Aggressiveness, and Fungicide Sensitivity of Phytophthora spp. Associated with Soybean in Illinois. Plant Disease. doi: 10.1094/PDIS-07-22-1551-RE
Chandra S, Choudhary M, Bagaria PK, et al. (2022) Progress and prospectus in genetics and genomics of Phytophthora root and stem rot resistance in soybean (Glycine max L.). Front Genet. 13: 939182. doi: 10.3389/fgene.2022.939182
Chen L, Zhang X, Wang W, et al. (2017) Network and role analysis of autophagy in Phytophthora sojae. Nature Scientific Reports 7, Article number: 1879. doi: 10.1038/s41598-017-01988-7
Chen X, Wang Y (2017) Phytophthora sojae. In: Wan F, Jiang M, Zhan A (Eds.) Biological Invasions and Its Management in China. Invading Nature – Springer Series in Invasion Ecology, vol 13. Springer, Singapore. pp. 199-223. doi: 10.1007/978-981-10-3427-5_15
Chen H, Fang Y, Song W, et al. (2023) The SET domain protein PsKMT3 regulates histone H3K36 trimethylation and modulates effector gene expression in the soybean pathogen Phytophthora sojae. Mol Plant Pathol. doi: 10.1111/mpp.13301
Cheng Y, Zhang H, Zhu W, et al. (2022) Ferroptosis induced by the biocontrol agent Pythium oligandrum enhances soybean resistance to Phytophthora sojae. Environ Microbiol. doi: 10.1111/1462-2920.16248
Chepsergon J, Motaung TE, Moleleki LN (2021) «Core» RxLR effectors in phytopathogenic oomycetes: A promising way to breeding for durable resistance in plants? Virulence 12(1): 1921-1935. doi: 10.1080/21505594.2021.1948277
Chowdhury RN, Tande C, Byamukama E (2021) Common Phytophthora sojae Pathotypes Occurring in South Dakota. Plant Health Progress. doi: 10.1094/PHP-02-21-0039-FI
de Ronne M, Santhanam P, Cinget B, et al. (2021) Mapping of partial resistance to Phytophthora sojae in soybean PIs using whole-genome sequencing reveals a major QTL. Plant Genome: e20184. doi: 10.1002/tpg2.20184
Dai TT, et al. (2012) Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 334: 27–34. doi: 10.1111/j.1574-6968.2012.02619.x
Deb D, Anderson RG, How-Yew-Kin T, et al. (2018) Conserved RxLR Effectors From Oomycetes Hyaloperonospora arabidopsidis and Phytophthora sojae Suppress PAMP- and Effector-Triggered Immunity in Diverse Plants. Molecular Plant-Microbe Interactions 31(3): 374-385. doi: 10.1094/MPMI-07-17-0169-FI
Dong S, Qutob D, Tedman-Jones J, et al. (2009) The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. PLoS One 4:e5556. doi: 10.1371/journal.pone.0005556
Dong S, Yu D, Cui L, et al. (2011) Sequence Variants of the Phytophthora sojae RXLR Effector Avr3a/5 Are Differentially Recognized by Rps3a and Rps5 in Soybean. PLoS ONE6(7): e20172. doi: 10.1371/journal.pone.0020172
Dorrance AE, Schmitthenner AF (2000) New Sources of Resistance to Phytophthora sojae in the Soybean Plant Introductions. Plant Disease 84(12): 1303-1308. doi: 10.1094/PDIS.2000.84.12.1303
Dorrance AE, Mills D, Robertson AE, et al. (2007). Phytophthora root and stem rot of soybean. The Plant Health Instructor. Reviewed 2012. doi: 10.1094/PHI-I-2007-0830-07
Dorrance AE (2018) Management of Phytophthora sojae of soybean: a review and future perspectives. Canadian Journal of Plant Pathology (accepted). doi: 10.1080/07060661.2018.1445127
Dorrance AE, Vargas A, Navarro-Acevedo K, et al. (2024) Picarbutrazox effectiveness added to a seed treatment mixture for management of Oomycetes that impact soybean in Ohio. Plant Dis. doi: 10.1094/PDIS-06-23-1223-RE
Dou DL, Kale SD, Wang X, et al. (2008) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20: 1930–1947. doi: 10.1105/tpc.107.056093
Du Q, Yang X, Zhang J, et al. (2018) Over-expression of the Pseudomonas syringae harpin-encoding gene hrpZm confers enhanced tolerance to Phytophthora root and stem rot in transgenic soybean. Transgenic Research 27(3): 277–288. doi: 10.1007/s11248-018-0071-4
Fan S, Dong L, Han D, et al. (2017) GmWRKY31 and GmHDL56 Enhances Resistance to Phytophthora sojae by Regulating Defense-Related Gene Expression in Soybean. Frontiers in Plant Science 8: 781. doi: 10.3389/fpls.2017.00781
Fan S, Zhang Z, Song Y, et al. (2022) CRISPR/Cas9-mediated targeted mutagenesis of GmTCP19L increasing susceptibility to Phytophthora sojae in soybean. PLoS One 17(6): e0267502. doi: 10.1371/journal.pone.0267502
Feng S, Shi J, Hu Y, et al. (2022) Genome-Wide Analysis of Soybean Lateral Organ Boundaries Domain Gene Family Reveals the Role in Phytophthora Root and Stem Rot. Front Plant Sci. 13: 865165. doi: 10.3389/fpls.2022.865165
Gao et al. (2021) Comparative analysis of Phytophthora genomes reveals oomycete pathogenesis in crops. Heliyon 7: e06317. doi: 10.1016/j.heliyon.2021.e06317
Gao RF, Wang JY, Liu KW, et al. (2021) Comparative analysis of Phytophthora genomes data. Data Brief. 39: 107663. doi: 10.1016/j.dib.2021.107663
Gao H, Jiang L, Du B, et al. (2022) GmMKK4-activated GmMPK6 stimulates GmERF113 to trigger resistance to Phytophthora sojae in soybean. Plant J. doi: 10.1111/tpj.15809
Giachero ML, Declerck S, Marquez N (2022) Phytophthora Root Rot: Importance of the Disease, Current and Novel Methods of Control. Agronomy 12(3): 610. doi: 10.3390/agronomy12030610
Grams N, Ospina-Giraldo M (2019) Increased expression of Phytophthora sojae genes encoding membrane-degrading enzymes appears to suggest an early onset of necrotrophy during Glycine max infection. Fungal Genetics and Biology 133: 103268. doi: 10.1016/j.fgb.2019.103268
Grijalba PE, Gally ME (2015) Virulence of Phytophthora sojae in the Pampeana Subregion of Argentina from 1998 to 2004. Journal of Phytopathology 163(9): 723–730. doi: 10.1111/jph.12369
Grijalba PE, Martínez MC, Guillin E (2020) Pathotype and SSR variation in Phytophthora sojae from the Argentinean Pampas. Journal of Phytopathology 168: 228– 243. doi: 10.1111/jph.12885
Grijalba PE, Ridao del CA, Guillin E, et al. (2020) Pathogenic diversity of Phytophthora sojae in the southeast of the Province of Buenos Aires. Tropical Plant Pathology 45: 397–401. doi: 10.1007/s40858-020-00364-7
Grijalba PE, Ridao Ad, Steciow MM (2021) Oomycetes species associated with soybean in Buenos Aires Province (Argentina). Phytoparasitica. doi: 10.1007/s12600-021-00921-z
Grijalba PE (2021) SOJA Oomycetes de suelo en Argentina. Orientación Gráfica Editora. Ciudad Autónoma de Buenos Aires. 72p. ISBN 978-987-1922-45-1.
Gui X, Zhang P, Wang D, et al. (2022) Phytophthora effector PSR1 hijacks the host pre-mRNA splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell 34: 3443–3459. doi: 10.1093/plcell/koac176
Guo M, Li B, Xiang Q, et al. (2021) Phosphite translocation in soybean and mechanisms of Phytophthora sojae inhibition. Pesticide Biochemistry and Physiology 172: 104757. doi: 10.1016/j.pestbp.2020.104757
Hai NTT, Cuong ND, Quyen NT, et al. (2021) Facile Synthesis of Carboxymethyl Cellulose Coated Core/Shell SiO2@Cu Nanoparticles and Their Antifungal Activity against Phytophthora capsici. Polymers (Basel) 13(6): 888. doi: 10.3390/polym13060888
Hale B, Ratnayake S, Flory A, et al. (2023) Gene regulatory network inference in soybean upon infection by Phytophthora sojae. PLoS ONE 18(7): e0287590. doi: 10.1371/journal.pone.0287590
, (2023) An updated assessment of the soybean–Phytophthora sojae pathosystem. Plant Pathology 00: 1– 18. doi: 10.1111/ppa.13713
Han Q, Feng H, Zhao H, et al. (2013) Effect of a benzothiadiazole on inducing resistance of soybean to Phytophthora sojae. Protoplasma 250: 471–481. doi: 10.1007/s00709-012-0430-6
Han X, Shen D, Xiong Q, et al. (2021) The plant beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl Environ Microbiol.: AEM0160121. doi: 10.1128/AEM.01601-21
Hebb LW, Bradley CA, Mideros SX, et al. (2021) Pathotype Complexity and Genetic Characterization of Phytophthora sojae Populations in Illinois, Indiana, Kentucky, and Ohio. Phytopathology. doi: 10.1094/PHYTO-12-20-0561-R
Hebb LM, Bradley CA, Telenko DEP, et al. (2023) Isolates of Phytophthora sansomeana Display a Range of Aggressiveness on Soybean Seedlings. Plant Health Progress 24: 171-179. doi: 10.1094/PHP-08-22-0075-RS
Helliwell EE, Lafayette P, Kronmiller BN, et al. (2022) Transgenic Soybeans Expressing Phosphatidylinositol-3-Phosphate-Binding Proteins Show Enhanced Resistance Against the Oomycete Pathogen Phytophthora sojae. Front Microbiol. 13: 923281. doi: 10.3389/fmicb.2022.923281
Hosseini B, Voegele RT, Link TI (2023) Diagnosis of Soybean Diseases Caused by Fungal and Oomycete Pathogens: Existing Methods and New Developments. J Fungi (Basel) 9(5):587. doi: 10.3390/jof9050587
Hou X, He Z, Che Z, et al. (2023) Molecular mechanisms of Phytophthora sojae avirulence effectors escaping host recognition. Front Microbiol. 13: 1111774. doi: 10.3389/fmicb.2022.1111774
Hu Y, Xin XF (2023) A sweet story from Phytophthora-soybean interaction. Trends Microbiol.: S0966-842X(23)00265-2. doi: 10.1016/j.tim.2023.09.004
Hunter S, Williams N, McDougal R, et al. (2018) Evidence for rapid adaptive evolution of tolerance to chemical treatments in Phytophthora species and its practical implications. PLoS One 13(12): e0208961. doi: 10.1371/journal.pone.0208961
Jiang B, Cheng Y, Cai Z, et al. (2020) Fine mapping of a Phytophthora-resistance locus RpsGZ in soybean using genotyping-by-sequencing. BMC Genomics 21: 280. doi: 10.1186/s12864-020-6668-z
Karhoff S, Vargas-Garcia C, Lee S, et al. (2022) Identification of Candidate Genes for a Major Quantitative Disease Resistance Locus From Soybean PI 427105B for Resistance to Phytophthora sojae. Front. Plant Sci. 13: 893652. doi: 10.3389/fpls.2022.893652
Kasteel M, Ketelaar T, Govers F (2023) Fatal attraction: How Phytophthora zoospores find their host. Semin Cell Dev Biol. 148-149: 13-21. doi: 10.1016/j.semcdb.2023.01.014
Kato F, Ando Y, Tanaka A, et al. (2022) Synthesis of aglycones, structure-activity relationships, and mode of action of lycosides as inhibitors of the asexual reproduction of Phytophthora. Biosci Biotechnol Biochem.: zbac179. doi: 10.1093/bbb/zbac179
Khatri P, Wally O, Rajcan I, Dhaubhadel S (2022) Comprehensive Analysis of Cytochrome P450 Monooxygenases Reveals Insight Into Their Role in Partial Resistance Against Phytophthora sojae in Soybean. Front Plant Sci. 13: 862314. doi: 10.3389/fpls.2022.862314
Kronmiller BA, Feau N, Shen D, et al. (2023) Comparative Genomic Analysis of 31 Phytophthora Genomes Reveals Genome Plasticity and Horizontal Gene Transfer. Mol Plant Microbe Interact. 36(1): 26-46. doi: 10.1094/MPMI-06-22-0133-R
Lannon K (2010) Phytophthora sojae. NC State University
Lebreton A, Labbé C, De Ronne M, et al. (2018) Development of a Simple Hydroponic Assay to Study Vertical and Horizontal Resistance of Soybean and Pathotypes of Phytophthora sojae. Plant Disease 102(1): 114-123. doi: 10.1094/PDIS-04-17-0586-RE
Lee S, Rouf Mian MA, Sneller CH, et al. (2014) Joint linkage QTL analyses for partial resistance to Phytophthora sojae in soybean using six nested inbred populations with heterogeneous conditions. Theoretical and Applied Genetics 127(2): 429–444. doi: 10.1007/s00122-013-2229-z
Li Q, Zhang M, Shen D, et al. (2016) A Phytophthora sojae effector PsCRN63 forms homo-/hetero-dimers to suppress plant immunity via an inverted association manner. Nature Scientific Reports 6: 26951. doi: 10.1038/srep26951
Li N, Zhao M, Liu T, et al. (2017) A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora sojae. Frontiers in Plant Science 8: 1185. doi: 10.3389/fpls.2017.01185
Li Soybean ZINC FINGER PROTEIN03 targets two SUPEROXIDE DISMUTASE1s and confers resistance to Phytophthora sojae. Plant Physiology 192: 633–647. doi: 10.1093/plphys/kiad083
Lin F, Zhao M, Baumann DD, et al. (2014) Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genomics 10: 15:18. doi: 10.1186/1471-2164-15-18
Lin X, Torres Ascurra YC, Fillianti H, et al. (2023) Recognition of Pep-13/25 MAMPs of Phytophthora localizes to an RLK locus in Solanum microdontum. Front Plant Sci. 13: 1037030. doi: 10.3389/fpls.2022.1037030
Liu H, An T, Zhao Y, et al. (2022) Benzoxazines in the root exudates responsible for nonhost disease resistance of maize to Phytophthora sojae. Phytopathology. doi: 10.1094/PHYTO-12-21-0508-R
Liu X, Li C, Chen Y, et al. (2022) Untargeted lipidomics reveals lipid metabolism disorders induced by oxathiapiprolin in Phytophthora sojae. Pest Manag Sci. doi: 10.1002/ps.7334
López-Casallas M, Guevara-Castro YA, Pisco-Ortíz YC, et al. (2020) Pudrición de raíces y tallo de la soya por Phytophthora sojae Kaufm. & Gerd. en la altillanura plana del departamento del Meta. Summa Phytopathologica 46(2): 113-120. doi: 10.1590/0100-5405/230356
Lu X, Yang Z, Song W, et al. (2022) The Phytophthora sojae effector PsFYVE1 modulates immunity-related gene expression by targeting host RZ-1A protein. Plant Physiol. : kiac552. doi: 10.1093/plphys/kiac552
Lygin AV, Zernova OV, Hill CB, et al. (2013) Glyceollin is an important component of soybean plant defense against Phytophthora sojae and Macrophomina phaseolina. Phytopathology 103(10): 984-94. doi: 10.1094/PHYTO-12-12-0328-R
Ma Z, Zhu L, Song T, et al. (2017) A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science (online). doi: 10.1126/science.aai7919
Madina MH, Santhanam P, Asselin Y, et al. (2022) Progress and Challenges in Elucidating the Functional Role of Effectors in the Soybean-Phytophthora sojae Interaction. J Fungi (Basel). 9(1): 12. doi: 10.3390/jof9010012
Matthiesen RL, Abeysekara NS, Ruiz-Rojas JJ, et al. (2016) A method for combining isolates of Phytophthora sojae to screen for novel sources of resistance to Phytophthora stem and root rot in soybean. Plant Disease 100: 1424-1428. doi: 10.1094/PDIS-08-15-0916-RE
Matthiesen RL, Schmidt C, Garnica VC (2021) Comparison of Phytophthora sojae Populations in Iowa and Nebraska to Identify Effective Rps Genes for Phytophthora Stem and Root Rot Management. Plant Health Progress. doi: 10.1094/PHP-02-21-0016-FI
Matthiesen RL, Schmidt C, Robertson AE (2021) Comparison of Baiting Techniques for Recovering Phytophthora sojae from Soybean Fields in Iowa. Plant Health Progress. doi: 10.1094/PHP-02-21-0040-FI
McCoy AG, Byrne AM, Jacobs JL, et al. (2022) Oomicide treated soybean seeds reduce early season stand loss to Phytophthora sojae. Crop Protection 157: 105984. doi: 10.1016/j.cropro.2022.105984
McCoy AG, Belanger RR, Bradley CA, et al. (2023) A global-temporal analysis on Phytophthora sojae resistance-gene efficacy. Nat Commun 14: 6043. doi: 10.1038/s41467-023-41321-7
Million CR, Wijeratne S, Karhoff S, et al. (2023) Molecular mechanisms underpinning quantitative resistance to Phytophthora sojae in Glycine max using a systems genomics approach. Front Plant Sci. 14: 1277585. doi: 10.3389/fpls.2023.1277585
Na R, Yu D, Qutob D, et al. (2013) Deletion of the Phytophthora sojae avirulence gene Avr1d causes gain of virulence on Rsp1d. Mol. Plant Microbe Interact. 26: 969–976. doi: 10.1094/MPMI-02-13-0036-R
Navarro-Acevedo KA, Wijeratne S, Culman SW, et al. (2021) The Effect of Incubation Temperature on the Species Composition of Phytophthora, Phytopythium, and Pythium Communities Associated with Soybean. Phytobiomes Journal 5: 133-144. doi: 10.1094/PBIOMES-01-20-0016-R
Neupane K, Ghimire B, Baysal-Gurel F (2022) Efficacy and Timing of Application of Fungicides, Biofungicides, Host-Plant Defense Inducers, and Fertilizer to Control Phytophthora Root Rot of Flowering Dogwoods in Simulated Flooding Conditions in Container Production. Plant Disease. doi: 10.1094/PDIS-02-22-0437-RE
Panda A, Sen D, Ghosh A, et al. (2018) EumicrobeDBLite: a lightweight genomic resource and analytic platform for draft oomycete genomes. Molecular Plant Pathology 19: 227–237. doi: 10.1111/mpp.12505
Pei Y, Si J, Navet N, et al. (2022) Two typical acyl-CoA-binding proteins (ACBPs) are required for the asexual development and virulence of Phytophthora sojae. Fungal Genet Biol. :103695. doi: 10.1016/j.fgb.2022.103695
Pei Y, Ji P, Si J, et al. (2023) A Phytophthora receptor-like kinase regulates oospore development and can activate pattern-triggered plant immunity. Nat Commun. 14(1): 4593. doi: 10.1038/s41467-023-40171-7
Qin J, Song Q, Shi A, et al. (2017) Genome-wide association mapping of resistance to Phytophthora sojae in a soybean [Glycine max (L.) Merr.] germplasm panel from maturity groups IV and V. PLoS ONE12(9): e0184613. doi: 10.1371/journal.pone.0184613
Qiu M, Li Y, Ye W, et al. (2021) A CRISPR/Cas9‐mediated in situ complementation method for Phytophthora sojae mutants. Molecular Plant Pathology 22: 373– 381. doi: 10.1111/mpp.13028
Qiu X, Kong L, Chen H, et al. (2022) The Phytophthora sojae nuclear effector PsAvh110 targets a host transcriptional complex to modulate plant immunity. Plant Cell: koac300. doi: 10.1093/plcell/koac300
Qiu M, Tian M, Yong S, et al. (2023) Phase-specific transcriptional patterns of the oomycete pathogen Phytophthora sojae unravel genes essential for asexual development and pathogenic processes. PLoS Pathog. 19(3): e1011256. doi: 10.1371/journal.ppat.1011256
, , , et al. (2024) Mining oomycete proteomes for phosphatome leads to the identification of specific expanded phosphatases in oomycetes. Molecular Plant Pathology 25: e13425. doi: 10.1111/mpp.13425
Qutob D, Tedman-Jones J, Dong S, et al. (2009) Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS ONE 4: e5066. doi: 10.1371/journal.pone.0005066
Rasoolizadeh A, Labbé C, Sonah H. et al. (2018) Silicon protects soybean plants against Phytophthora sojae by interfering with effector-receptor expression. BMC Plant Biol 18, 97. doi: 10.1186/s12870-018-1312-7
Rolling W, Lake R, Dorrance AE, McHale LK (2020) Genome-wide association analyses of quantitative disease resistance in diverse sets of soybean [Glycine max (L.) Merr.] plant introductions. PLoS ONE 15(3): e0227710. doi: 10.1371/journal.pone.0227710
Sahoo DK, Abeysekara NS, Cianzio SR, et al. (2017) A Novel Phytophthora sojae Resistance Rps12 Gene Mapped to a Genomic Region That Contains Several Rps Genes. PLoS ONE12(1): e0169950. doi: 10.1371/journal.pone.0169950
Sahoo DK, Das A, Huang X, et al. (2021) Tightly linked Rps12 and Rps13 genes provide broad-spectrum Phytophthora resistance in soybean. Sci Rep 11: 16907. doi: 10.1038/s41598-021-96425-1
, , (2023) A rapid molecular diagnostic tool to discriminate alleles of avirulence genes and haplotypes of Phytophthora sojae using high-resolution melting analysis. Molecular Plant Pathology 00: 1–11. doi: 10.1111/mpp.13406
Santos G, Marchioro VS, Meira D, et al. (2023) Phytophthora root characterization in different phenological stages of soybean. Physiological and Molecular Plant Pathology 126: 102039. doi: 10.1016/j.pmpp.2023.102039
Sawake MM, Moharil MP, Ingle YV, et al. (2022) Management of Phytophthora parasitica causing gummosis in citrus using biogenic copper oxide nanoparticles. J Appl Microbiol. doi: 10.1111/jam.15472
Schneider R, Rolling W, Song Q, et al. (2016) Genome-wide association mapping of partial resistance to Phytophthora sojae in soybean plant introductions from the Republic of Korea. BMC Genomics 17: 607. doi: 10.1186/s12864-016-2918-5
Sepiol CJ, Yu J and Dhaubhadel S (2017) Genome-Wide Identification of Chalcone Reductase Gene Family in Soybean: Insight into Root-Specific GmCHRs and Phytophthora sojae Resistance. Frontiers in Plant Science 8: 2073. doi: 10.3389/fpls.2017.02073
Shan W, Cao M, Leung D, Tyler BM (2004) The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17: 394–403. doi: 10.1094/MPMI.2004.17.4.394
Shrestha SD, Chapman P, Zhang Y, Gijzen M (2016) Strain Specific Factors Control Effector Gene Silencing in Phytophthora sojae. PLoS ONE 11(3): e0150530. doi: 10.1371/journal.pone.0150530
Si J, Pei Y, Shen D, et al. (2021) Phytophthora sojae leucine-rich repeat receptor-like kinases: diverse and essential roles in development and pathogenicity. iScience 24(7): 102725. doi: 10.1016/j.isci.2021.102725
Si J, Pei Y, Ji P, et al. (2021) PsGRASP, a Golgi Reassembly Stacking Protein in Phytophthora sojae, Is Required for Mycelial Growth, Stress Responses, and Plant Infection. Front Microbiol. 12: 702632. doi: 10.3389/fmicb.2021.702632
Situ J, Xi P, Lin L, et al. (2022) Signal and regulatory mechanisms involved in spore development of Phytophthora and Peronophythora. Front Microbiol. 13: 984672. doi: 10.3389/fmicb.2022.984672
Stewart S, Wickramasinghe D, Dorrance AE, Robertson AE (2011) Comparison of three microsatellite analysis methods for detecting genetic diversity in Phytophthora sojae (Stramenopila: Oomycete). Biotechnology Letters 33: 2217. doi: 10.1007/s10529-011-0682-9
Stewart S, Abeysekara N, Robertson AE (2014) Pathotype and genetic shifts in a population of Phytophthora sojae under soybean cultivar rotation. Plant Disease 98: 614-624. doi: 10.1094/PDIS-05-13-0575-RE
Stewart S, Robertson AE, Wickramasinghe D, et al. (2016) Population Structure Among and Within Iowa, Missouri, Ohio, and South Dakota Populations of Phytophthora sojae. Plant Disease 100(2): 367-379. doi: 10.1094/PDIS-04-15-0437-RE
Sugano S, Sugimoto T, Takatsuji H, Jiang C‐J (2013) Induction of resistance to Phytophthora sojae in soyabean (Glycine max) by salicylic acid and ethylene. Plant Pathol, 62: 1048-1056. doi: 10.1111/ppa.12011
Sugimoto T, Watanabe K, Yoshida S, et al. (2010) Field application of calcium to reduce Phytophthora stem rot of soybean, and calcium distribution in plants. Plant Dis. 94: 812–819. doi: 10.1094/PDIS-94-7-0812
Sugimoto T, Kato M, Yoshida S, et al. (2012) Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breeding Scince 61(5): 511–522. doi: 10.1270/jsbbs.61.511
Sun Y, Wang Y, Zhang X, et al. (2022) Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature. doi: 10.1038/s41586-022-05214-x
Tian M, Zhao L, Li S, et al. (2016) Pathotypes and metalaxyl sensitivity of Phytophthora sojae and their distribution in Heilongjiang, China 2011–2015. Journal of General Plant Pathology 82(3): 132–141. doi: 10.1007/s10327-016-0654-y
Torto TA, Li S, Styer A, et al. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13: 1675–1685. doi: 10.1101/gr.910003
Tremblay V, McLaren DL, Kim YM, et al. (2021) Molecular assessment of pathotype diversity of Phytophthora sojae in Canada highlights declining sources of resistance in soybean. Plant Disease. doi: 10.1094/PDIS-04-21-0762-RE
Tsypurskaya EV, Nikolaeva TN, Lapshin PV, et al. (2022) Response of Transgenic Potato Plants Expressing Heterologous Genes of ∆9- or ∆12-Acyl-lipid Desaturases to Phytophthora infestans Infection. Plants (Basel) 11(3): 288. doi: 10.3390/plants11030288
Tyler BM (2006) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Molecular Plant Pathology 8(1): 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Tyler BM, Tripathy S, Zhang X, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266. doi: 10.1126/science.1128796
Tyler BM (2009) Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cellular Microbiology 11(1): 13–20. doi: 10.1111/j.1462-5822.2008.01240.x
, , , et al. (2021) Mining germplasm panels and phenotypic datasets to identify loci for resistance to Phytophthora sojae in soybean. Plant Genome 14: e20063. doi: 10.1002/tpg2.20063
, , , (2010) GPR11, a putative seven-transmembrane G protein-coupled receptor, controls zoospore development and virulence of Phytophthora sojae. Eukaryot. Cell, 9: 242–250. doi: 10.1128/EC.00265-09
Wang Q, Han C, Ferreira AO, et al. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23: 2064–2086. doi: 10.1105/tpc.111.086082
Wang Y, Ye W, Wang Y (2018) Genome‐wide identification of long non‐coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae. Molecular Plant Pathology 19: 2177-2186. doi: 10.1111/mpp.12692
Wang Y, Wang Y (2018) Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 46: 7–13. doi: 10.1016/j.mib.2018.01.008
Wang W, Jiao F (2019) Effectors of Phytophthora pathogens are powerful weapons for manipulating host immunity. Planta 250: 413–425. doi: 10.1007/s00425-019-03219-x
Wang M, Du Y, Ling C, et al. (2021) Design, Synthesis and Antifungal/Anti‐Oomycete Activity of Pyrazolyl Oxime Ethers as Novel Potential Succinate Dehydrogenase Inhibitors. Pest Manag Sci. Accepted Author Manuscript. doi: 10.1002/ps.6418
Wang W, Chen L, Fengler K, et al. (2021) A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat Commun. 12(1): 6263. doi: 10.1038/s41467-021-26554-8
Wang H, Guo B, Yang B, et al. (2021) An atypical Phytophthora sojae RxLR effector manipulates host vesicle trafficking to promote infection. PLoS Pathog 17(11): e1010104. doi: 10.1371/journal.ppat.1010104
Wang Z, Lv X, Wang R, et al. (2022) Use of oxathiapiprolin for controlling soybean root rot caused by Phytophthora sojae: Efficacy and mechanism of action. Pest Manag Sci. doi: 10.1002/ps.7207
Wang Z, Ke Q, Tao K, et al. (2022) Activity and Point Mutation G699V in PcoORP1 Confer Resistance to Oxathiapiprolin in Phytophthora colocasiae Field Isolates. J Agric Food Chem. doi: 10.1021/acs.jafc.2c06707
Wang S, Zhang X, Zhang Z, et al. (2023) Fusarium-produced vitamin B6 promotes the evasion of soybean resistance by Phytophthora sojae. J Integr Plant Biol. doi: 10.1111/jipb.13505
Wilcox JR, Martin SK (1998) Soybean Genotypes Resistant to Phytophthora sojae and Compensation for Yield Losses of Susceptible Isolines. Plant Disease 82(3): 303-306. doi: 10.1094/PDIS.1998.82.3.303
Winkworth RC, Neal G, Ogas RA, et al. (2022) Comparative analyses of complete Peronosporaceae (Oomycota) mitogenome sequences – insights into structural evolution and phylogeny. Genome Biol Evol. evac049. doi: 10.1093/gbe/evac049
Xue Z, Wang W, Shen J, et al. (2021) A Patched-Like Protein PsPTL Is Not Essential for the Growth and Response to Various Stresses in Phytophthora sojae. Front Microbiol. 12: 673784. doi: 10.3389/fmicb.2021.673784
Yan H, Nelson B Jr. (2019) Adaptation of Phytophthora sojae to Rps Resistance Genes over the Past Two Decades in North Dakota. Plant Health Progress 20: 88-93. doi: 10.1094/PHP-10-18-0062-RS
Yang B, Wang Y, Guo B, et al. (2019) The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytologist 222: 425-437. doi: 10.1111/nph.15581
Yang J, Zheng S, Wang X, et al. (2020) Identification of Resistance Genes to Phytophthora sojae in Domestic Soybean Cultivars from China Using Particle Bombardment. Plant Dis. 104(7): 1888-1893. doi: 10.1094/PDIS-10-19-2201-RE
Yang X, Jiang X, Yan W, et al. (2021) The Mevalonate Pathway Is Important for Growth, Spore Production, and the Virulence of Phytophthora sojae. Front Microbiol. 12: 772994. doi: 10.3389/fmicb.2021.772994
Yin W, Dong S, Zhai L, et al. (2013) The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol. Plant Microbe Interact. 26: 958–968. doi: 10.1094/MPMI-02-13-0035-R
Yu X, Tang J, Wang Q, et al. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytologist 196(1): 247–260. doi: 10.1111/j.1469-8137.2012.04241.x
Yu S-F, Wang C-L, Hu Y-F, et al. (2022) Biocontrol of Three Severe Diseases in Soybean. Agriculture 12(9): 1391. doi: 10.3390/agriculture12091391
Zhang M, Rajput NA, Shen D, et al. (2015) A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana. Nature Scientific Reports 5, Article number: 10837 . doi: 10.1038/srep10837
Zhang X, Zhai C, Hua C, et al. (2016) A Gα-binding protein is required for chemotaxis. Molecular Plant Pathology 17: 272-285. doi: 10.1111/mpp.12279
Zhang C, Wang X, Zhang F, et al. (2017) Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Nature Scientific Reports 7, Article number: 7242. doi: 10.1038/s41598-017-07832-2
Zhang X, Liu B, Zou F, et al. (2019) Whole Genome Re-sequencing Reveals Natural Variation and Adaptive Evolution of Phytophthora sojae. Front. Microbiol. 10:2792. doi: 10.3389/fmicb.2019.02792
Zhang M, Liu Y, Li Z (2021) The bZIP transcription factor GmbZIP15 facilitates resistance against Sclerotinia sclerotiorum and Phytophthora sojae infection in soybean. iScience 24(6): 102642. doi: 10.1016/j.isci.2021.102642
Zhang B, Zhang Z, Yong S, et al. (2022) An oomycete-specific leucine-rich repeat-containing protein is involved in zoospore flagellum development in Phytophthora sojae. Phytopathology. doi: 10.1094/PHYTO-12-21-0523-R
Zhang Z, Lin L, Chen H, et al. (2022) ATAC-seq reveals the landscape of open chromatin and cis-regulatory elements in the Phytophthora sojae genome. Mol Plant Microbe Interact. doi: 10.1094/MPMI-11-21-0291-TA
Zhang F, Chen S, Cui T, et al. (2023) Novel function of the PsDMAP1 protein in regulating the growth and pathogenicity of Phytophthora sojae. Int J Biol Macromol.: 127198. doi: 10.1016/j.ijbiomac.2023.127198
Zhang Y, Zhang Z, Chen Y, et al. (2023) Protein Kinase A Regulatory Subunit is Required for Normal Growth, Zoosporogenesis, and Pathogenicity in Phytophthora sojae. Res Microbiol.:104152. doi: 10.1016/j.resmic.2023.104152
Zhao Y, Yang B, Xu H, et al. (2022) The Phytophthora effector Avh94 manipulates host jasmonic acid signaling to promote infection. J Integr Plant Biol. doi: 10.1111/jipb.13358
Zhong C, Sun S, Yao L, et al. (2018) Fine Mapping and Identification of a Novel Phytophthora Root Rot Resistance Locus RpsZS18 on Chromosome 2 in Soybean. Frontiers in Plant Science 9: 44. doi: 10.3389/fpls.2018.00044